DID YOU SEE THE GOOGLE ADSENSE ADS ON THE RIGHT?
1) INTRODUCTION
 Try a Google search on "intelligent bacteria", bacterial signalling, "quorum sensing", "bacterial neural networks" and even "bacterial evolutive learning" or "bacterial multi-cellularity"... You'll come up with papers by people like Bonnie Bassler, Eshel Ben Jacob, Klaas Hellingwerf, Claudio Aguilar, James Shapiro, and now, Saeed Tavazoie. What do these works all have in common?
Try a Google search on "intelligent bacteria", bacterial signalling, "quorum sensing", "bacterial neural networks" and even "bacterial evolutive learning" or "bacterial multi-cellularity"... You'll come up with papers by people like Bonnie Bassler, Eshel Ben Jacob, Klaas Hellingwerf, Claudio Aguilar, James Shapiro, and now, Saeed Tavazoie. What do these works all have in common?They promote a view of bacterial colonies as super-organisms having sophisticated, computing behaviors and even some form of logical computation and elementary thinking (in the sense for instance of a loose artificial neural network). They even start to speak about learning in bacterial communities.
This is what this post is all about: in which sense can we say that a bacterial colony is a sort of elementary proto-brain, able to compute and learn?
Why are we interested in this? In the preceding posts, you met what I call" Andrones" and "Bactorgs".
Andrones are interconnected sets of real neurons, living in a culture medium on top of a multi-electrode array connected to a computer. They learn to do what real neural networks do : to exchange electrical and chemical signals to produce quite complex behaviors (drawing something pleasant for us on a piece of paper, controlling the flight of a model plane, doing logical computations and so on...). These applications do really exist. Of course, they are still a bit rudimentary and need a lot of progress. Even so, the very fact of their existence is a testimony to the ingenuity of the researchers who designed them. It is one of the true adventures of modern experimental science and engineering. Just finding how to make these in vitro neurons to live, interconnect and learn is a first rate accomplishment
Yet, on a more theoretical viewpoint, Andrones are not so astonishing... They learn to compute. Well, after all, that's what neurons have evolved to do. They develop some form of rudimentary intelligence and are happy afterwards forever ... no big theoretical breakthrough...
Bactorgs are completely different. First they do not exist or are not yet really acknowledged. In "WE-SHARE" (as you will remember, this is the title of my novel), they are bacterial multi-species communities developing also a rudimentary form of thinking and learning (or perhaps, after all, not so rudimentary...).
Think twice,... "thinking bacteria"? I'll adopt a very limited definition of what thinking is but still, that's a big step to take. As you know, I want, all my premises to be very realistic scientifically speaking. So, I have better to document this point very carefully.
The first point I'll discuss is that, under certain conditions, a bacterial colony behaves, not as a collection of separated individuals, but as a coordinated whole, i.e. an integrated organism. This is clearly a prerequisite to act as a protobrain.
2) A BACTERIAL COLONY AS A MULTICELLULAR ORGANISM
It all starts with James Shapiro who, in a 1988 Scientific American paper, proposed that a bacterial colony was not to be seen as a collection of individual cells but as an integrated organism having its own unity and emergent behaviors not deducible from the comportments of the isolated bacteria. (see Sci. Am; 1988, 256; 82 - 89).
Read this paper, it is a must. Its idea was initially received with much skepticism...; usual is n'it? But paper after paper, many researchers elaborated upon and ten years later, Shapiro was able to put together a wonderful review paper on the progresses made during the first decade of live of the concept of "bacterial multicellular organism". It was found that several species were developing colonies acting as multicellular organisms having coordinated behaviors: development of structured colonies, swarming, metabolic cooperation and much more (see "Thinking about bacterial populations as multicellular organisms, Ann. Rev. of Microbiology, 1998, 104, 52-81).
It was also found that bacteria benefit from this multicellular organization by using cellular division of labor, accessing resources that cannot be effectively utilized by single cells and optimizing population survival by differentiating into distinct cell types.
Fast forward ten more years and, today, in 2008, bacterial multi-cellularity has become a very important way of thinking, an emerging paradigm. It has been found that cell to cell communication mechanisms (a.k.a. quorum sensing) is present in virtually all species. It has also been found that bacterial colonies grown under usual laboratory conditions (what we call now "domesticated cultures") present much less intercellular features than so called" wild colonies", grown in nature or in conditions emulating nature. In retrospect, this is no wonder, usual practice in microbiology does all it can to isolate cells and subcolonies. No wonder they loose intercellular communication and coordination.
The picture hereafter is taken (with permission) from a paper by Claudio Aguilar, Hera Vlamakis, Richard Losick and Roberto Kolter (from Harvard) (Thinking about bacillus subtilis as a multicellular organism published in Curr. Opinion Microbiol. 2007, 10(6): 638-643).
Claudio Aguilar ----------------------- Roberto Kolter
Their paper is a tribute to Shapiro and presents a recent summary of the field. On the left, you see three wild colonies showing clearly intricate structures. On the right, you see the corresponding "domesticated" cultures showing much less structure (they are mainly simple blobs...). If you want to study bacterial organisms, take a walk on the wild side...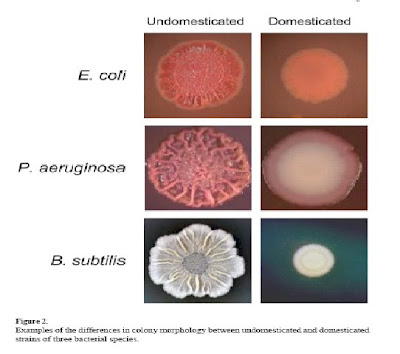
So, wild colonies are multicellular and organized. If you need supplementary arguments think about Eshel Ben Jacob's work which we discussed in a previous post... wonderful multicellular structures. It is then normal to think that there are some computations done in the wild colonies to synchronize and maintaintheir structures and affect different roles to bacteria at different places.
Our second step is now to suggest that these multicellular organisms do not only compute but do it almost as neural networks. Ben Jacob, as we have seen, clearly suggests it. However, his arguments are indirect. Can we say something about the cellular or genetic mechanisms used by a single bacterium in these multicellular
organisms to do their bit of computation?
I will now discuss the work of Klaas Hellingwerf, the guy who has proposed to take seriously the analogy between ANNs and bacterial signalling networks.
A WARNING: Below, I will suppose that you have at least some general notions on artificial neural networks. Later, I will post a short primer on neural networks. Here I will just discuss how bacterial networks fit or do not fit the framework of neural networks. For more details, see later.
4) NEURAL NETWORKS ANALOGUES IN A SINGLE BACTERIUM?
 As I promised you before, we will go now one step further in the direction of thinking bacteria. Meet Klaas Hellingwerf from
As I promised you before, we will go now one step further in the direction of thinking bacteria. Meet Klaas Hellingwerf from
Klaas speaks somewhat metaphorically (or perhaps not so metaphorically) about "bacterial neural networks" and in 2002, he organized an European EURESCO conference on this theme in Obernai (France). The conference, attended by about 200 people elicited a wide interest in bacterial computations (interconnected phenomena of signaling, behavior and development) which has now become a big theme in microbiology with surveys published in some major journals.
See for instance an EMBO report by Susan Golden (Texas A&M) on this conference entitled "Think like a bacterium"... (EMBO reports Vol 4, N°1, 2003, pages 15-17).
See also another report published in "Molecular microbiology (2003, 47(2), 583-593" by Judith Armitage, Professor of biology at
These reports show that some form of crude "bacterial thinking" (I mean "thinking as it is done in an artificial neural network" - see Rumelhart PDPs or Mc Culloch and Pitts) , is now a serious scientific subject and no longer exclusively the stuff of science fiction.
What is thus Hellingwerf's argument? I will summarize it from one of his papers entitled "Bacterial observations: a rudimentary form of intelligence" (Trends in microbiol., 13, 4, 2005, 152 - 158).
He starts by saying: "Until very recently, bacteria were considered too small to be little more than bags of enzymes unable to realize complex processes like signal transduction, association, gene expression, response to various stimuli, intra and extra-cellular communication. This is no longer so. We know now that even a single bacterium has many regulating mechanisms and can use them to express genetically the required chemical components for each of the above processes at specific times and places."
Then his argument goes a little bit like this: "Most notably, signal transduction can take an (extra) cellular signal S of a chemical or physical nature (e.g. light or perhaps electricity or electromagnetic waves) and convert it into a different form called response R (for instance a transduction of light into a given concentration of some protein which, then, can affect gene expression or enzyme activity and lead to specific behaviors (e.g. chemotaxis, phototaxis, swimming)." The figure below is modified from his paper, see above, and gives a schematic representation of a typical S-R system.
Legend:
What is this? Just a genetic embodiment of the familiar S-R (stimulus response) generic model of biological signal processing! Several genetic S-R systems may be present in a single bacterium, all different but operating in parallel on various signals to produce various responses. These mechanisms form what we may call a "genetic network of signal processing".
Neuronal networks are also signal processing networks. Klaas proposes that the S-R networks of bacteria may abstractly be considered as functional equivalents of simple neuronal networks (i.e. accomplish the same kind of abstract computational algorithms but of course with different mechanisms and signals). To be considered as functionally equivalent to a neuronal network, Klaas says that our bacterial network must satisfy four properties:
- There must be many parallel S-R mechanisms (pathways) and these pathways must be branched (e.g. an individual S-R mechanism may have several inputs coming from the environment or from other S-R mechanisms and several outputs going into effectors or to other S-R mechanisms.). Neural networks do this because signals have multiple pathways and do many computations in parallel. A bacterium does this since it has several messaging pathways like the one illustrated above in parallel. Remark that traditional computers are not parallel and thus do not fit the paradigm. It is possible of course to simulate a parallel network on a serial computer but not in real time.
- These S-R pathways must execute logical operations. Computational nodes must combine the signals from two or more previous elements, compute an output depending on all the incoming signals and pass the result to another node. The result must be able to be represented approximatively by a mathematical or logical function (E.g.: and, or, not...). Klaas does argue that bacteria do this because their signaling systems combine inputs from different sources. Non linearity is essential.
- There must be some auto amplification mechanisms (feedback). This is a very important property which means that a computing node (e.g. an enzyme) acts as a non linear function of an input. The reason for this requirement is that it is very important for a neuron to have an output which is a non linear (for instance, a sigmoid) function of a combination of its inputs. The logical, classification and learning properties of an artificial neuron depends critically on this sigmoid-like output (treshold behavior, back propagation).
Bacteria may do that quite easily. Suppose that a signal is used to generate a small amount of a given chemical. If this chemical is auto-catalysed (as it is the case for many genetic expressions). The response chemical will use cellular resources to synthesize itself more and more leading to an enormous increase in its concentration (the sigmoid response). Thus as soon as a threshold is reached, the autocatalysis mechanism sets in and the sigmoid response is reached. If it is very strong and quick, it may even be seen as an all or none response (Boolean response, see René Thomas and kinetic logic in Google).
- There must be some significant amount of cross-talk between mechanisms. This is where the difficulty lies for a single bacterium. This means that parallel chain reactions of signal response must exchange signals so that the way one chain operates change the ways the other run. Again this is essential in artificial neural networks if we want them to have interesting behaviors like distributed processing and coding, associative memory, generalization, graceful degradation or complex classification. Klaas says that there is some scarce evidence for crosstalk among signaling pathways in a single bacterium. Yet, today, the operative word is "scarce".
Based on existing detailed experimental work, he then suggests that the bacteria Sacharomyces Cerevisiae presents these four features. However, he insists that evidence for crosstalk is still quite small.
The figure hereafter shows his view of such a S-R network in a single bacterium. Each circle in the upper membrane shows a molecule receiving an input S (a chemical, a light signal, an electrical signal, an electromagnetic radiation and as we will see later even a sound wave) .
Then the red arrows show the internal pathway from the various S to various R (blue nodes are intermediate chemicals. Responses are gene expression, membrane processes activation, flagellar movement and the like. You can see the multi-input, multi-output feature. The green circles show the auto amplification of some chemicals; the blue interrupted lines show the putative cross talk interactions still to investigate. Clearly this is a neural network analogue.
Klaas ends up by proposing that, if we can make some experimental progress to demonstrate cross talk in a single bacterium, we will be entitled to see signal processing in a single bacterium as an analogue to a simple neural network. So, speaking metaphorically a bacterium will do neural computations, i.e. "think" if we adopt a crude operational definition of thinking as “doing what artificial neurons networks do”.
4) A NETWORK VIEW BASED, NOT ON A SINGLE BACTERIUM BUT ON A COLONY
I personally do not believe that seeing a single bacterium as analogue to a neural network is really mandatory. I think that what might be important is that the bacterium can process information in one or several coupled S-R networks doing logical operations with or without crosstalk. That seems to be experimentally demonstrated. Then, a single bacterium is more like a simplified neuron, what I would like to call a “proto-neuron” or a set of proto-neurons in parallel (each S-R mechanism being one) without much crosstalk.
Then consider a set of several millions (or billions) of bacteria (proto-neurons) and suppose they exchange signals between them. If the signals have some specificity (What John Holland calls tagged signals: a signal carries with it a part which tells which receivers can receive it, so there is communication specificity). Then various bacteria are sensitive to different signals and process them differently. You may thus consider that the signals diffuse in the medium but that a given bacteria receives only some of them. It selects its signals. If, in your mind, you link then by an arrow the bacteria which are able to exchange a signal, you see a network developing. The connections are not hardwired like those between neurons (axons, dendrites) but much more labile and dependent on who emits what and who receives what. Here is your cross talk, outside the bacteria... at the community level. The result looks much like a simplified collective brain (a proto-collective brain) analog to those described for ants or termites. What I envision is thus this: one or several proto-neurons per bacterium; exchange of many different signals between neurons (ex: tagged by intensity or by chemical nature or by association of different signals); receptivity of different bacteria or sets of bacteria to different signals (and thus development of an implicit network with cross talk at the level of a set of sets of bacteria.
The result looks much like a simplified collective brain (a proto-collective brain) analog to those described for ants or termites. What I envision is thus this: one or several proto-neurons per bacterium; exchange of many different signals between neurons (ex: tagged by intensity or by chemical nature or by association of different signals); receptivity of different bacteria or sets of bacteria to different signals (and thus development of an implicit network with cross talk at the level of a set of sets of bacteria.
There is no reason why such a collective brain made of tens of billions of bacteria could not be, on its own evolutionary time scale, as powerful as the collective brain of a colony of ants. Of course, I have replaced Klaas's cross talk in a bacterium by cross talk between bacteria. So, I have now to look if this hypothesis makes sense.
It is time to meet somebody else who study just that: networks of bacteria talking together in a common language (i.e. exchanging signals). You will meet Bonnie Bassler, from Princeton ( I realize that until now, most of my posts have been a bit superficial and introductory. They also have been experimentally oriented and not theoretical. I'll have to equilibrate that somewhat. Do not forget: as a great engineer once said (Th. von Karman), " There is nothing more practical than a good theory".
I realize that until now, most of my posts have been a bit superficial and introductory. They also have been experimentally oriented and not theoretical. I'll have to equilibrate that somewhat. Do not forget: as a great engineer once said (Th. von Karman), " There is nothing more practical than a good theory".




1 comment:
Very informative. Specially the part about collective brains and the hypothesis that they could be at work in bacterial colonies. I like the bactorgs
Post a Comment